Synthetic genetic circuits in plants could be the next technological horizon in plant breeding, showcasing potential for precise patterned control over expression. Nevertheless, uncertainty in metabolic environments prevents robust scaling of traditional genetic circuits for agricultural use, and studies show that a deterministic system is at odds with biological randomness. We analyze the necessary requirements for assuring Boolean logic gate sequences can function in unpredictable intracellular conditions, followed by interpreted pathways by which a mathematical representation of probabilistic circuits can be translated to biological implementation. This pathway is utilized through translation of a probabilistic circuit model presented by Pervaiz that works through a series of bits; each composed of a weighted matrix that reads inputs from the environment and a random number generator that takes the matrix as bias and outputs a positive or negative signal. The weighted matrix can be biologically represented as the regulatory elements that affect transcription near promotors, allowing for an electrical bit to biological bit translation that can be refined through tuning using invertible logic prediction of the input to output relationship of a genetic response. Failsafe mechanisms should be introduced, possibly through the use of self-eliminating CRISPR-Cas9, dosage compensation, or cybernetic modeling (where CRISPR is clustered regularly interspaced short palindromic repeats and Cas9 is clustered regularly interspaced short palindromic repeat-associated protein 9). These safety measures are needed for all biological circuits, and their implementation is needed alongside work with this specific model. With applied responses to external factors, these circuits could allow fine-tuning of organism adaptation to stress while providing a framework for faster complex expression design in the field.
Published at: https://acsess.onlinelibrary.wiley.com/doi/10.1002/tpg2.20525
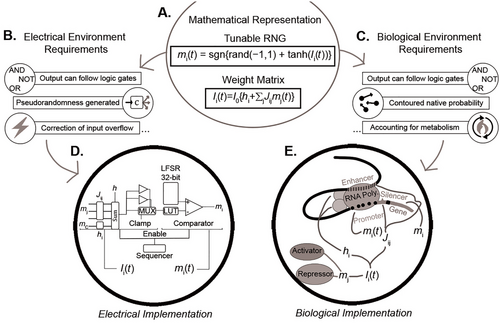
Figure: Pathway of probabilistic circuits from math to method. The Pervaiz mathematical representation provides the tunable random number generator (RNG) function and the weight matrix. The environmental requirements determine which processes this representation must undergo to be physically interpreted. The electrical environment requires the circuit to generate its own randomness and correct for input outside the expected range. The biological pathway requires interpretation of the native Brownian motion-based probability and accounting for metabolic constraints. Both pathways also need an interpretation to be capable of handling Boolean logic operations. These operations are modular enough to allow the circuit to represent complex functions. The electrical implementation includes the weight matrix through wired interconnections in a column-wise arrangement. It also clamps overflowing values at this stage using a multimixer (MUX). The RNG is implemented with a 32-linear feedback shift register (LFSR). This register is a chain of bistable circuit latches connected sequentially, fed information about previous states. The resulting random number is compared to a tanh activation function implemented using a lookup table (LUT). The biological implementation uses a genetic promotor as its tunable RNG, interpreting the randomness present. Regulatory elements effectively serve as the weight matrix. Individual transcription factors may also serve as functional inputs or local bias tuners based on metabolic conditions. Both implementations serve the same function. However, each does so in reference to the local needs surrounding them. This is true of any pathway that stems from the mathematical representation: it must be able to acknowledge and be informed by its setting.

